Software#
A full list of software that can perform the local structure modeling for total scattering data will be presented in this page. Also, links to relevant tutorials are provided. There are two main categories of software for total scattering data analysis – those based on unit-cell approach and those based on supercell approach. The former one is similar to the Rietveld fitting for Bragg diffraction data, where one has a unit cell defined for describing the underlying crystal structure and in practice one has a few structural parameters such as lattice constant, atomic motif positions, atomic displacement parameters, etc. to fit against the experimental total scattering data. For such an approach, one is assuming that the local structural correlation is represented by the unit cell, i.e., periodic bounday condition is assumed beyond the unit cell length scale. One can view such a unit cell as an average representative of the overall local scale structure aspect. However, such an average is different from the average unit cell as seen by the Bragg diffraction. The difference between the average as seen these two techniques is illustrated in the picture as follows,
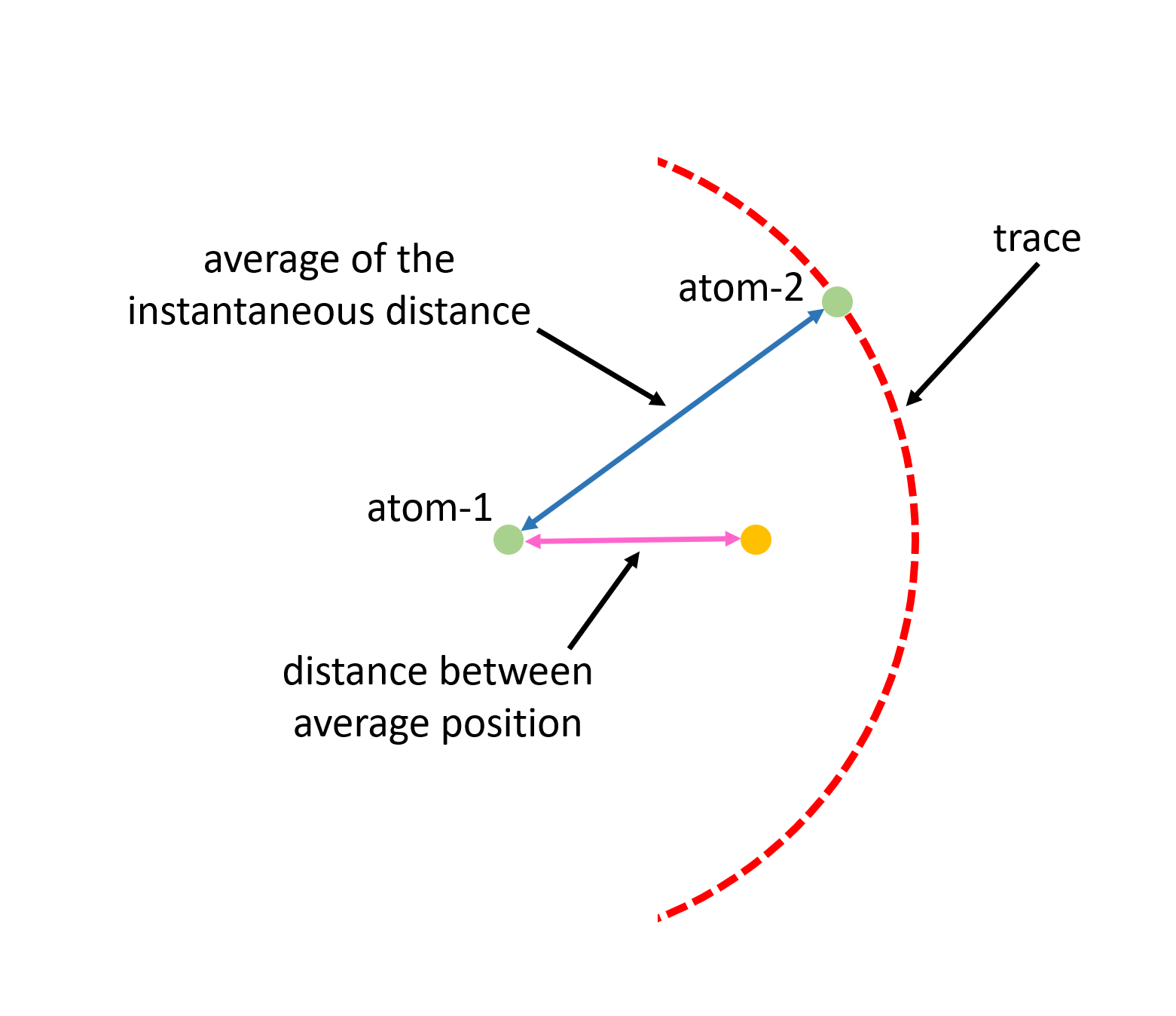
Suppose we have two atoms with atom-1 fixed in the middle and atom-2 moving around atom-1 along a circular trace (the red dashed circle in the image), the local probe (e.g., with total scattering) sees the instantaneous distance between the two atoms and thus the average distance as seen by the local probe is the average of the instantaneous distances, which is, in this case, just the radius of the circle. However, from the average structure perspective, as seen by the Bragg diffraction, the average of the distance between atom-1 and atom-2 corresponds to the distance between the average position of the two atoms. In this demo case, average position of atom-1 is the center of the circle and the average position of atom-2 is represented by the orange dot in the image. Therefore, the average distance as seen by the Bragg diffraction is that represented by the pink arrow in the image.
Another typical approach for fitting the total scattering data is based on the supercell approach, using the reverse Monte Carlo method. In such an approach, one starts from a unit cell and expand it to a supercell. Afterwards, on then remove the symmetry constraint so the structure model then becomes a bix box (i.e., supercell). Then one adjust the model to match the data until a good agreement is achieved, and thus the supercell approach is considered as a data driven approach.
On top of the driving force from the experiment data, one could also add in more constraint based on physical or chemical consideration of the underlying system, such as distacne window, bond valence sum, etc.
Different software has its own way of deciding a ‘good’ agreement, but the fundamental idea is universal where one needs to define a threshold to be used as the criterion of convergence, i.e., good agreement with experimental data.
Due the the large parameter space in the supercell approach, overfitting may become an issue. Typically, one may need to comprehensively inspect the outcome from the supercell data fitting. Or, a commonly used method is to repeat the same set of fitting multiple times to accumulate statistics. Some advanced methods concerning the overfitting issue are yet to be developed, e.g., given entropy consideration.
Unit-cell based#
Diffpy-CMI#
DiffPy-CMI is a flexible library of Python modules for robust modeling of structures in crystals, nano-, and amorphous materials. The package provides a fitting framework for combining multiple experimental inputs. Diffpy-CMI is available on Linux and Macintosh machines.
Link to software: https://www.diffpy.org/products/diffpycmi/index.html
Tutorial
DISCUS#
DISCUS package consists of four sections, including those for generating structure model, calculation and fitting of scattering pattern, Monte Carlo simulation and data plotting.
Link to software: https://tproffen.github.io/DiffuseCode/
Tutorial
PDFgui#
PDFgui is a convenient and easy to use graphical front end for the PDFfit2 refinement program. It is capable of full-profile fitting of the atomic pair distribution function (PDF) derived from x-ray or neutron diffraction data and comes with built in graphical and structure visualization capabilities.
Link to software: https://www.diffpy.org/products/pdfgui.html
We have been seeing quite a few issues that people are having in installing PDFgui using conda. Here I am posting the installation procedure that I found to be working robustly, using mamba.
Install
mamba, following the instruction here.For Unix-like platforms (macOS, Linux, & WSL), the shell initialization will be done after the installation finishes and we just need to follow the command line prompt. For
Windows, after the installation, we need to find and launchMiniforge PromptinStart. Then on theMiniforge Promptcommand line, we typemamba shell init. This is for the purpose of initializing the Windows terminal (e.g.,CMD) so that themambacommand can be executed from terminal other thanMiniforge Prompt.It is recommended to install the full version of
mambausing the installer here. Otherwise,Miniforge Promptwill not be installed.After all the previous steps (installation and initialization), we want to open a terminal and type
mambato see whether it is working.Then proceed to install
PDFgui,mamba create -n diffpy.pdfgui_env diffpy.pdfgui mamba activate diffpy.pdfgui_env python -c "import diffpy.pdfgui; print(diffpy.pdfgui.__version__)"
Topas#
Topas is a commercial package for analyzing Bragg diffraction data and meanwhile it has the capability of refining the total scattering data in both real and reciprocal space.
Link to software: http://www.topas-academic.net/
Tutorial
Supercell based#
Dissolve#
A simulation tool for the interrogation of neutron scattering data, typically total neutron scattering data. It builds on the techniques established in the Empirical Potential Structure Refinement (EPSR) method by Soper.
Link to software: https://github.com/disorderedmaterials/dissolve
Tutorial
Empirical Potential Structure Refinement (EPSR)#
EPSR is a computational technique addressing the problem of calculating a three-dimensional structure which exploits the information contained in diffraction data. It is based around an atomistic Monte Carlo simulation of a system, correcting the employed pair interaction potentials by comparison with the diffraction data sets.
Link to software: https://www.isis.stfc.ac.uk/Pages/Empirical-Potential-Structure-Refinement.aspx
Tutorial
fullRMC#
fullRMC is an RMC engine for refining the local structure model with the supercell approach written in Python (the underlying calculation engine is in Cython).
Link to software: https://bachiraoun.github.io/fullrmc/index.html
Tutorial
RMCProfile#
RMCProfile was built from the original RMCA code of McGreevy & Pusztai to determine the local structure of crystalline materials while still being capable of analyzing disordered systems. The current version of RMCProfile results from a collaboration between scientists at ISIS facility (UK), Spallation Neutron Source (SNS at Oak Ridge National Laboratory, US), University of Cambridge (UK), University of Oxford (UK), Queen Mary University of London (QMUL, UK) and National Institute of Standards and Technology (NIST, US). It is now possible to fit many data types simultaneously (Neutron & X-ray total scattering & the Bragg profile, EXAFS, single crystal diffuse scattering) and use a range of constraints to produce atomic models that are consistent with all the available data. In this way we are progressing the effort to develop a ‘complex modeling’ approach to elucidate structural details of materials that are the key to their exploitable functional properties.
Link to software: https://rmcprofile.ornl.gov/
Tutorial
For magnetic structure modeling#
Since neutrons carry spins, it can be scattered by magnetic moments in magnetic materials. Analogous to the nucleus total scattering pattern, we also have the magnetic total scattering pattern and in practice it will come together with the nucleus total scattering signal. For the data analysis, similar to its nucleus counterpart, one can either follow the unit-cell or supercell based approach. Besides, one can also choose to analyze the magnetic total scattering data in either reciprocal space, without Fourier transform, or real space, with Fourier transform to obtain the so-called magnetic pair distribution function (mPDF). One is recommended to read about Ref. [et al., 2019] for a summary diagram about the available approaches for magnetic total scattering data analysis. Here, we will put down some useful tools for the magnetic total scattering data analysis.
diffpy.mpdf#
The diffpy.mpdf package provides a convenient method for computing the magnetic PDF (mPDF) from magnetic structures and performing fits to neutron total scattering data.
Link to software: https://www.diffpy.org/products/mPDF.html
Tutorial
RMCProfile#
Details about the RMCProfile package has already been covered above. Apart from the capability of modeling the nucleus total scattering data, the RMCProfile package can also model the magnetic total scattering data, through a combined refinement for both the nucleus and magnetic total scattering in the same scope.
Link to software: https://rmcprofile.ornl.gov/
Tutorial
Spinvert#
Spinvert is a program for refinement of for refinement of atomistic models to powder magnetic diffuse scattering data for frustrated magnets, spin glasses, and other magnetically disordered materials.
Link to software: https://www.icloud.com/iclouddrive/0ECSZy25kk__w7cYyI3fRgCyA#spinvert_18Mar19
Tutorial
Spinteract#
Spinteract is a program for refinement of magnetic interaction parameters to magnetic diffuse scattering data collected on powder and single-crystal samples.
Link to software: https://www.icloud.com/iclouddrive/0ECSZy25kk__w7cYyI3fRgCyA#spinvert_18Mar19
Tutorial
